Sneaking up on tiny crystals with electron diffraction
Researchers accelerate protein structure determination with electrons
Understanding the structure of proteins, the building blocks of life, is essential to obtain insight into their biological function. Due to their minute size and extreme fragility, these structures are enormously difficult to determine. Acquiring data of sufficient resolution requires immense doses of high energy X-ray radiation, which unfortunately irrevocably damage the proteins principally being investigated.
Now researchers from the MPSD and DESY in Hamburg have developed an inventive new method which avoids these pitfalls and uses accessible, cost-effective technology. Their work describing the new method has now been published in Nature Communications.

For decades, researchers from many fields such as physics, biology, and biochemistry have poured their creativity into circumventing the radiation-damage conundrum. Current approaches include the use of extremely short and intense X-ray flashes at facilities such as the new European X-ray Free Electron Laser (EuXFEL) in Hamburg, which can take well-exposed images of proteins before literally making them explode.
While this method has been spectacularly successful in obtaining high resolution protein structures, generating X-ray beams of the required brightness necessitates the use of large and expensive particle accelerators. A highly effective alternative, which is intensively practiced at the Center for Structual Systems Biology (CSSB) in Hamburg, for example, is to forgo X-rays altogether, and use electron beams instead, which are gentler to the delicate biomolecules and easier to generate.
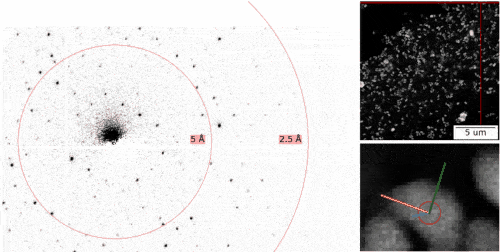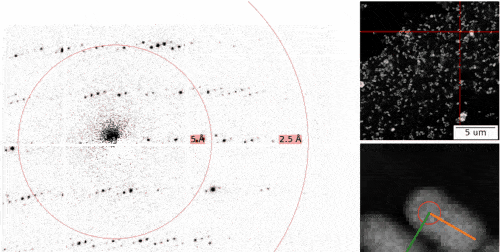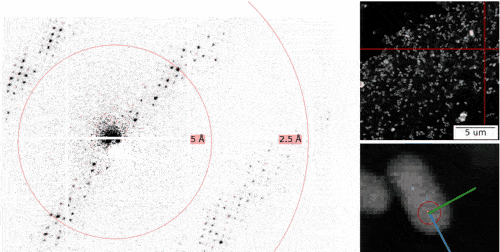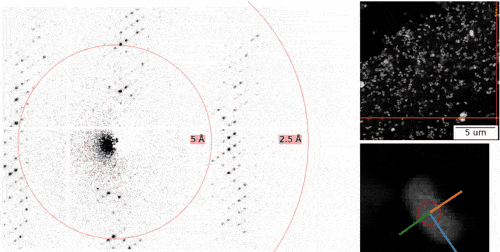
The MPSD/DESY research team at the Center for Free-Electron Laser Science (CFEL), which also includes scientists from the Hamburg Cluster of Excellence CUI : Advanced Imaging of Matter, has ingeniously combined such methodologies with big data computing and recent improvements in camera technology and managed to obtain high resolution protein structures from relatively easily obtainable nano-crystals. To achieve this, they have developed a technique called serial electron diffraction by adapting experimental methods already known in the X-ray crystallography community for sequentially acquiring and processing diffraction patterns from thousands of crystals.
Instead of deploying a billion-Euro instrument such as the EuXFEL, they simply distributed these crystals on a thin carbon film and inserted them into a transmission electron microscope, a ubiquitously available device. The electron beam is made to hop from one nano-crystal to the next in order to acquire diffraction data. Apart from material savings on often rare and costly samples, utilizing nano-crystals means that researchers no longer have to grow large protein crystals as required by older (X-ray) methods - an undertaking that often proves prohibitively difficult.
To work around the damage caused by the electron beam, instead of only taking a single photograph, a short movie is recorded using a high-speed camera while the electron beam is resting on each crystal. In the movie, one can literally watch the proteins in the crystal “melt away” – however, there is enough information in this diffraction-during-destruction movie to reconstruct data nearly as if there was no damage at all. This procedure is repeated for thousands of nano-crystals, and within a few hours, using specialized software developed at DESY, the massive amount of data is converted into a high-resolution protein structure.
In addition to proteins and other biomolecules, serial electron diffraction is also applicable to many classes of novel functional materials, such as perovskites and metal-organic frameworks – all of them promising candidates for future applications in solar cells and hydrogen storage. The research team is excited about the ease of use of this innovative technique, with its low equipment requirements and wide applicability. They anticipate that it will spread from the MPSD to labs worldwide.













